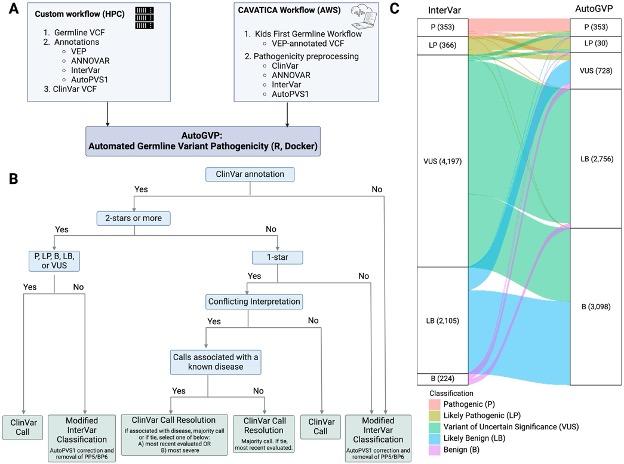
Automated Germline Variant Pathogenicity (AutoGVP) integrates ClinVar variant annotation with a modified InterVar classification approach, based on American College of Medical Genetics-Association for Molecular Pathology (ACMG-AMP) guidelines, to output germline variant classification. Since AutoGVP input only requires a VCF file, it can facilitate large-scale, clinically focused classification of germline sequence variants.
Using automated germline classification reduces hands-on time and allows for reproducibility of variant classification.
Website
AutoGVP is an open source, dockerized workflow implemented in R and freely available on GitHub.
Access AutoGVP enabled for Biowulf (on GitHub).
Support
Contact: Jung Kim
Reference
Kim J et al. AutoGVP: a dockerized workflow integrating ClinVar and InterVar germline sequence variant classification. Bioinformatics. 2024.