Single Cell Atlas Characterizes Mechanisms of Lung Cancer Risk Variants
, by Jennifer K. Loukissas, M.P.P.
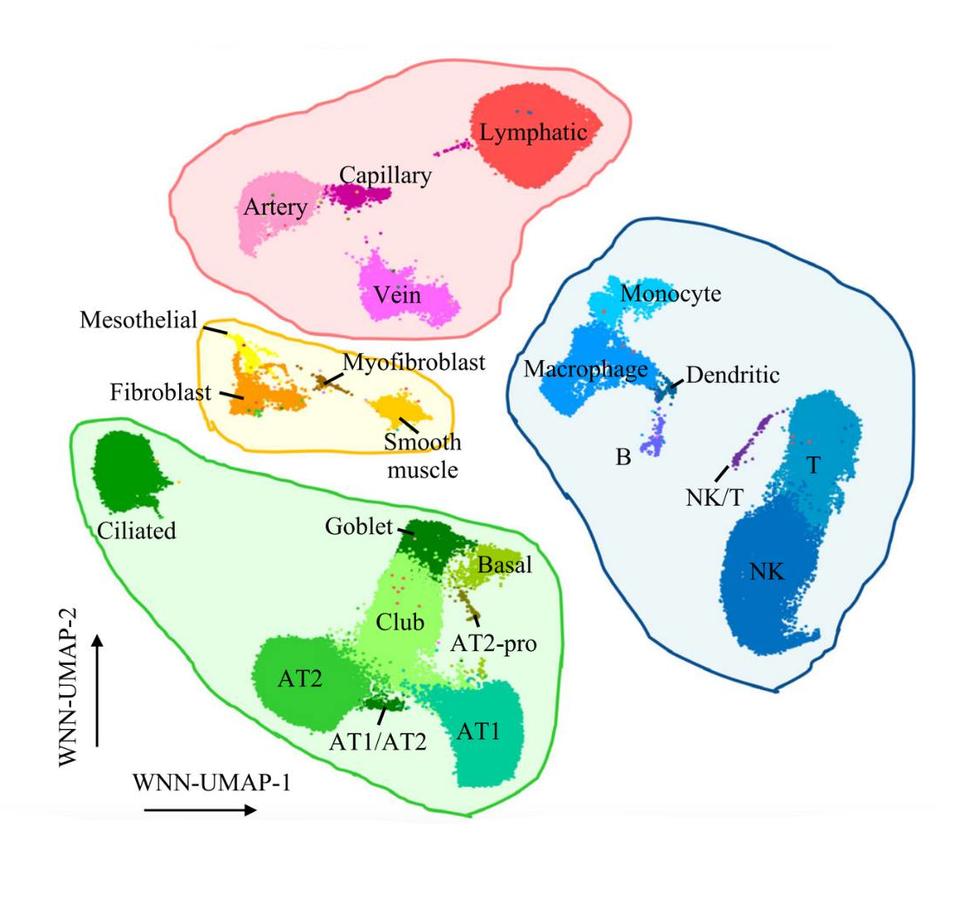
Multiple genetic variants are associated with lung cancer risk, but the underlying genetic mechanisms remain poorly understood, in part due to difficulties in detecting context-specific gene regulation by non-protein-coding variants. To address this challenge, Jiyeon Choi, Ph.D., M.S., Earl Stadtman investigator in the Laboratory of Translational Genomics, and her colleagues have evaluated the function of variants in specific cell types found in the lung, using single-cell multiomics assays, to jointly profile gene expression and chromatin accessibility (i.e., potentially active regions of genome). The researchers, led by Dr. Choi’s former postdoctoral fellow Erping Long, Ph.D., identified new lung cancer susceptibility genes from loci previously identified in genome-wide association studies (GWAS) and characterized molecular contexts where such variants are functional. Their findings were published September 12, 2024, in the journal Nature Communications.
The researchers used single-cell multiomics assays to look at genomic regions with potential function in lung cell types, revealing that most of the potentially functional genomic regions are highly cell-type-specific across the 23 different lung cell types. By directly overlaying lung cancer GWAS variants with these potentially functional regions, the researchers assessed how much of lung cancer risk variation is explained by different cell-type-specific regulatory regions of lung. Their results indicated that the GWAS-identified genes contribute to lung cancer risk the most through the potentially functional regions of epithelial (where most lung cancers arise) and immune cells.
By linking these cell-type-specific potentially functional regions with gene expression, the researchers discovered previously undetected target genes. Most GWAS hits were linked to multiple target genes through more than one cell-type-specific non-coding regions that have possible regulatory function. This result indicates that complex context-specific gene regulations by GWAS variants are very prevalent. These data will help researchers better understand when and where lung cancer-associated genetic variants are functional, adding to our understanding of lung tumorigenesis.
Although there have been many single-cell studies for lung tissues, this is the first study to jointly profile gene expression and chromatin accessibility. Given that many cell types important in lung cancer development constitute a small fraction of normal lung tissue and are often difficult to culture, this single-cell dataset could provide unique information to the research community. Download detailed raw and processed single-cell data through GEO GSE241468.
Reference
Long E et al. Context-aware single-cell multiomics approach identifies cell-type-specific lung cancer susceptibility genes. Nature Commun. 2024