Beyond the Signature: Exposing Mutational Patterns of Cancer
, by Justine E. Yu, Ph.D.
Mutational signature analysis is a powerful emerging tool that can be used to determine the underlying processes that contribute to tumor development and identify the populations that may benefit from tailored screening, treatment, and prevention strategies.
Uncovering a Tumor's Evolutionary Journey
A tumor’s genetic code is akin to a roadmap that scientists can use to track its evolutionary journey, specifically, the accumulation of somatic mutations that caused it to develop. Technologies to analyze genomic data from a collection of tumors can reveal distinct molecular or genetic patterns or mutational signatures. These patterns can be used to infer associations of internal processes, such as DNA repair deficiency, or external exposures, such as carcinogens (e.g., radiation, chemotherapy, tobacco), with a particular type of cancer. Mutational signature analysis is a powerful tool for natural history and etiologic studies and could identify high-risk populations that may benefit from tailored screening, treatment, and prevention strategies.
"Advances in DNA sequencing technology have enabled us to begin to address some of the important questions underlying the evolution of cancer, in part through comprehensive genomic analyses carried out in well-designed epidemiological studies," said Stephen J. Chanock, M.D., DCEG Director.
By sequencing the genome of both the tumor tissue and paired germline DNA, researchers can catalog the counts and types of mutations to define the genomic landscape of a tumor: single base substitutions, double base substitutions, small insertions and deletions, structural variants, and somatic copy number alterations. Novel computational analyses can decipher the mutation data and extract patterns that can be linked to endogenous processes or exposures. While numerous mutational signatures have been identified, for many, their causes are still unknown.
Surveying the Genomic Landscape
DCEG scientists seek to uncover the relationship between these unique signature patterns and the mutational mechanisms through cross-functional collaborations, by utilizing rich data resources, often the result of major international partnerships or consortia.
“Defining mutational signatures in different cancer types is paramount to understanding cancer etiology, uncovering prognostic and therapeutic biomarkers, and determining clinical strategies,” said Montserrat García-Closas, M.D., Dr.Ph.H., Deputy Director of DCEG, and Director and senior investigator of the Trans-Divisional Research Program (TDRP).
Endogenous Mutagenic Processes
Somatic Copy Number Alterations (The Sherlock-Lung Study)
While cigarette smoking is an established cause of lung cancer, 15-20% of lung cancer cases arise in people with no history of smoking; epidemiologists have long sought to understand this phenomenon. A recent genomic analysis of lung cancer in never smokers led by Tongwu Zhang, Ph.D., staff scientist, and Maria Teresa Landi, M.D., Ph.D., senior investigator, both in the Integrative Tumor Epidemiology Branch (ITEB), found that these tumors clustered in three subtypes based on somatic copy number alterations. Borrowing terminology from musical composition, the researchers defined the three molecular subtypes observed based on the amount of genomic ‘noise’ or mutations. The most common subtype—“piano”—had the fewest mutations, had stem-like properties, and were slow growing. The “mezzo-forte” subtype had specific amplifications and mutations in the growth factor receptor gene EGFR, which is commonly altered in lung cancer. The “forte” subtype had whole-genome doubling, which is also a common feature in lung tumors in smokers.
“We’re starting to distinguish cancer subtypes that could guide the development of more precise approaches for prevention and treatment,” said Dr. Landi. For example, the slow-growing piano could give clinicians a window of opportunity to detect these tumors earlier when they are easier to treat. In contrast, the mezzo-forte and forte have only a few major driver mutations, suggesting that these tumors could be identified by a single biopsy and benefit from already available targeted treatments, she said.
DNA Editing Through APOBEC Mutagenesis: Helpful and Harmful
APOBEC3 enzymes fight infections at the cellular level by mutating viral genetic material. Bin Zhu, Ph.D., tenure-track investigator in the Biostatistics Branch (BB) and Lisa Mirabello, Ph.D., M.S., senior investigator in the Clinical Genetics Branch, investigated the APOBEC3 response to human papillomavirus 16 (HPV 16) infection. APOBEC3 editing of the HPV 16 genome induced mutations that may reduce the viability of HPV 16, resulting in viral clearance, and may also contribute to the genetic diversity of HPV 16.
In the process of editing viral DNA, APOBEC3 can also unintentionally mutate the host DNA, causing damage that could contribute to the development of cancer. APOBEC-mediated DNA damage follows a specific pattern that has been observed in many tumor types. Abdul Rouf Banday, Ph.D., research fellow, and Ludmila Prokunina-Olsson, Ph.D., Chief and senior investigator, both of the Laboratory of Translational Genomics (LTG), published the first study linking the APOBEC mutational signature in bladder tumors with a germline variant in the APOBEC3 region on chromosome 22, identified in bladder cancer genome wide association studies (GWAS).
While APOBEC mutagenesis can contribute to tumor evolution and resistance to treatment, Drs. Banday, Prokunina-Olsson, and colleagues, found that increased APOBEC mutagenesis found in carriers of the risk allele of the bladder cancer-associated variant, was associated with improved survival of aggressive, muscle-invasive bladder cancer. “One explanation is that higher mutation load may improve the effectiveness of treatments that target tumor DNA. Tumor cells with more mutations are also more vulnerable to synthetic lethality due to mutation combinations, and they express more cell-surface neoantigens that can trigger the patient’s immune response, leading to elimination of the tumor cells,” said Dr. Prokunina-Olsson. Thus, the clinical significance of the APOBEC mutational signature may be tumor-type specific and warrants further studies on the biological mechanisms which regulate it.
Influence of the Microenvironment
While changes in the internal processes of tumor cells, like DNA editing, replication, and repair, are common sources of mutational signatures, the tumor microenvironment—comprised of the surrounding cells and tissues—may also play a role. Jill Koshiol, Ph.D., senior investigator in the Infections and Immunoepidemiology Branch (IIB), profiled the mutational landscape of gallbladder cancer and identified three subtypes, two associated with poorer survival. Further investigation into the molecular mechanisms of these survival differences found that the subtypes associated with poor survival had immunosuppressive microenvironments and inhibited immune function. These findings elucidate the influence of the tumor microenvironment on mutational signatures.
Exogenous Carcinogenic Exposures
Cigarette Smoking
DCEG investigators continue to provide insights into the molecular mechanisms that impact smoking-related carcinogenesis. A team of researchers, including Stella Koutros, Ph.D., M.P.H., tenure-track investigator in the Occupational and Environmental Epidemiology Branch, Michael Dean, Ph.D., senior investigator in LTG, and Dr. Prokunina-Olsson, and colleagues, used targeted sequencing of frequently mutated genes in bladder cancer to characterize the mutational signatures of bladder cancer. They observed two predominant single base substitution signatures in bladder tumors, APOBEC signature and ERCC2-specific mutational signature, were associated with cigarette smoking, and that there was heterogeneity in the relationship between smoking status. Specifically, they found the burden of ERCC2 signature mutations was higher in current smokers while the burden of APOBEC signature mutations was higher in former smokers. In both instances, they observed a strong association with smoking duration (the component most strongly associated with bladder cancer risk). These data quantify the contribution of smoking to mutational burden and suggest different signature enrichment among never, former, and current smokers.
Ionizing Radiation
In 1986, an explosion at the Chernobyl nuclear power plant in northern Ukraine exposed millions of individuals in the surrounding region to radioactive contaminants, resulting in increased number of papillary thyroid carcinoma (PTC) in children exposed to radioactive iodine (I-131). Lindsay M. Morton, Ph.D., Deputy Chief and senior investigator in the Radiation Epidemiology Branch, and colleagues, characterized the genomic landscape of radiation-induced cancer by exploring the effect of ionizing radiation on thyroid cancer risk in people exposed as children or in utero to I-131 released by the Chernobyl nuclear accident. The researchers found that DNA double-strand breaks are early events following radiation exposure that enable PTC growth, and the number of double-strand breaks are correlated with increasing radiation dose, especially for individuals exposed at younger ages.
Advancement of Mutational Signature Methodology
Despite advances in sequencing technology and computational methods, mutational signature analysis can present many technical challenges which DCEG scientists are actively working to improve, from techniques to preserve samples to statistical approaches to address missing data.
Semiparametric kernel independence test (SKIT)
While many signatures have been identified, the underlying processes that created them are still being discovered. Dr. Zhu and DongHyuk Lee, Ph.D., postdoctoral fellow in BB, develop methods to extract mutational signatures and characterize their etiologies in different studies across the Division. They recently demonstrated that if two signatures are associated, then they may also share correlated exposures (or the same exposure in special cases). That is, if the exposure of one signature is known, and the exposure of an associated signature is unknown, then it is likely that the unknown exposure is associated with the known exposure. However, investigators may encounter the problem of excess zero values in mutational signature data, where signatures are present in some patients but not others. Drs. Zhu and Lee designed a method to address this challenge—the semiparametric kernel independence test (SKIT). This approach models excess zeroes in the independence test to increase statistical power.
Optimizing the Use of Formalin-Fixed Paraffin-Embedded Tissues
Formalin-fixed paraffin-embedded (FFPE) processing is the standard method of preservation of tissue samples for pathological diagnosis or experimental research, but the fixation process can affect DNA quality. This presents many challenges in downstream applications such as molecular testing and genomic sequencing. Despite the significant shortcomings, comprehensive bioinformatic analyses integrated with visual inspection of the data can improve detection of somatic mutations. Alyssa Klein, M.S., Dr. Zhang, and other ITEB collaborators are working to develop a bioinformatic pipeline to optimize the results from FFPE-derived sequencing data by providing systematic and comprehensive mutation calling, filtering, evaluation, and visualization from DNA sequencing data derived from FFPE samples. A statistical method will be used to remove the formalin-induced artefacts based on extracted FFPE signatures.
Integrative mutational signature portal (MsigPortal)
As the number of mutational signatures with known etiology has increased from many different cancer genomic studies, there is critical need for a curated census of signatures as well as mutational signature-related data sharing. Drs. Zhang and Landi, in collaboration with the NCI Center for Biomedical Informatics and Information Technology, are developing the integrative mutational signatures portal (mSigPortal) which will enable users to comprehensively explore, visualize, and analyze the mutational signature related data (including mutational profiles, signatures, proposed etiology, tissue specificity, activity, and association) in cancer genomic studies. mSigPortal will allow users to analyze their own datasets as well as explore and analyze large, collected datasets from public cancer genomic studies, such as TCGA, Pan-Cancer Analysis of Whole Genomes, Breast Cancer 560 Whole Genome Sequences, and Sherlock-Lung. This portal will facilitate broad investigation of mutational signatures to elucidate different mutagenic processes involved in tumorigenesis.
Overcoming Low-Mutation Count Data
So far, applications of mutational signature analysis have been largely limited to whole-exome sequencing, which covers the protein-coding portion of an individual’s DNA, and whole-genome sequencing which covers the entire DNA. However, targeted gene sequencing panels, which only sequence selected regions of interest, are most commonly used in oncology clinics. Unlike whole-genome/whole-exome sequencing, these panels capture considerably smaller regions of the genome and therefore generate low-mutation count data. As a result, there is a need to develop new robust methods to detect mutational signatures from low-mutation count data.
The development of novel and robust methods to detect signatures from low-mutation count data is being undertaken by a collaboration involving Clara Bodelon, Ph.D., staff scientist in ITEB, Jonas S. Almeida, Ph.D., Chief Data Scientist, Jeya Balaji Balasubramanian, Ph.D., postdoctoral fellow, and Aaron Ge, summer intern, in TDRP. They have been developing and evaluating various machine learning models to detect signatures from low-mutation count data to enable the use of data from targeted sequencing panels to inform cancer prevention and treatment.
Ongoing and Future Initiatives
B-CAST: Stratifying Breast Cancer
The international Breast CAncer STratification (B-CAST) initiative, led by Dr. García-Closas and colleagues within the Breast Cancer Association Consortium (BCAC), is designed to analyze targeted sequencing panel data from 11,000 breast cancer tumors. These sequencing data will be integrated with automated digital pathology analyses of 15 markers in over 20,000 breast tumors in B-CAST, being co-led by Mustapha Abubakar, M.D., Ph.D., research fellow in ITEB, and colleagues, in collaboration with the Molecular and Digital Pathology Lab. This is the largest set of breast cancer tumors with sequencing data and detailed risk factor, genetic, and pathological data, and has the potential to provide important etiological and prognostic clues for breast cancer.
Environmental Exposures: Coal Combustion, Diesel Exhaust, and Perfluorooctanoic Acid
Qing Lan, M.D., Ph.D., M.P.H., senior investigator in OEEB, is leading efforts to study lung cancer in Xuanwei, China, where rates among never-smokers are among the highest in the world, primarily due to household air pollution from burning a highly carcinogenic type of “smoky” (i.e., bituminous) coal. To understand the impact of smoky coal on patterns of somatic mutations in lung tumors among never-smokers, Dr. Lan, in collaboration with Nathaniel Rothman, M.D., M.P.H., M.H.S., senior investigator in OEEB, Drs. Zhang, Landi, and Chanock, Huu Phuc Hoang, Ph.D., of ITEB, and Wei Hu, Ph.D., staff scientist in OEEB, are conducting the first whole genome sequencing study to compare mutational signatures in lung tumor samples from Xuanwei smoky coal users to residents of the same province that rely on a different fuel source.
Diesel exhaust has been linked to increased bladder cancer risk, but few studies have provided insight into diesel-related bladder carcinogenesis. Drs. Koutros and Dean are using genomic characterization of bladder cancer to explore the mutagenic signature patterns associated with nitro-polycyclic aromatic hydrocarbon exposures, which are markers of the potent mutagenic constituents of diesel exhaust, and are linking these with quantitative estimates of occupational exposure in the New England Bladder Cancer Study.
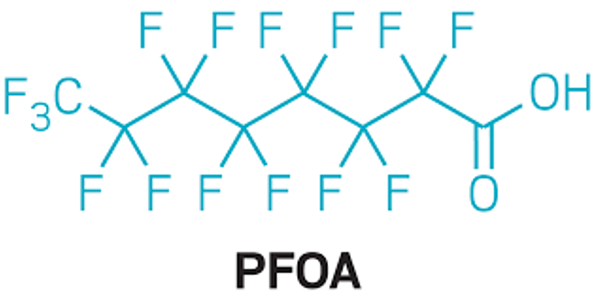
Perfluorooctanoic acid (PFOA) and other per- and polyfluoroalkyl substances (PFAS), are a diverse class of organic pollutants used in commercial and industrial applications, classified as a possible human carcinogen by the International Agency for Research on Cancer in 2014. A recent study led by Jonathan Hoffman, Ph.D., M.P.H., tenure-track investigator in OEEB, found that high serum concentrations of PFOA were associated with an increased risk of kidney cancer, adding to the mounting evidence of PFOA as a renal carcinogen. Mark Purdue, Ph.D., senior investigator in OEEB, and collaborators, are launching a new study within the Mutographs of Cancer Project (funded by Cancer Research UK with contributions from the NCI European Kidney Cancer Study) to explore the PFOA and other PFAS-related molecular mechanisms underlying renal carcinogenesis. Results from this study will provide critical insight as to the role of PFOA/PFAS in cancer etiology and may have important public health implications for the many individuals exposed to this ubiquitous and highly persistent chemical in the U.S. and worldwide.
Comprehensive Etiological Studies of Lung Cancer
Building on the success of the Sherlock-lung study, the ITEB investigators are looking ahead to other investigations of lung cancer in never-smokers. “We want to extend our studies to more diverse populations and explore factors beyond carcinogenic exposures, to environmental or lifestyle factors that indirectly initiate the endogenous processes that lead to cancer,” said Dr. Landi. To that end, she is working with Jiyeon Choi, Ph.D., M.S., Earl Stadtman tenure-track investigator in LTG, and Dr. Chongyi Chen in the NCI Center for Cancer Research, to explore whether inflammatory processes that promote tissue damage and regeneration with activation of stem cells contribute to tumor initiation. In addition, they will expand their study population to include a wide range of ancestries and racial and ethnic backgrounds. Drs. Lan and Rothman are conducting Sherlock-lung studies in other Asian populations, which will enable comparisons of tumor mutation and other molecular patterns within and between Asian and other populations and potentially identify new etiologic agents.
Dr. Lan leads additional studies of lung cancer in never-smokers that evaluate a wide range of exogenous exposures and endogenous processes, with a particular focus on East Asian populations. These include a large-scale GWAS with Jianxin Shi, Ph.D., senior investigator in BB, and Drs. Choi, Rothman and Chanock; studies of gene-environment interactions and risk prediction models with postdoctoral fellow Batel Blechter, Ph.D., and research fellow Jason Wong Sc.D., in OEEB; and nested case-control studies within the Asia Cohort Consortium of exposure and intermediate biomarkers including epigenome-wide methylation with Dr. Wong, postdoctoral fellow Charles Breeze, Ph.D., and staff scientist Mohamad L. Rahman, M.D., Sc.D., M.P.H., in OEEB. These studies will help inform lung cancer mutational signatures.
Jian Sang, Ph.D., postdoctoral fellow in ITEB and colleagues, are seeking to understand the complex processes between exposure time and intensity from tobacco smoking with the development of a mutational signature. Using the tobacco smoking associated mutational signature (i.e., cosmic mutational signature SBS4), in the Environment And Genetics in Lung cancer Etiology (EAGLE) study, they will measure time from exposure, and time from quitting smoking, to detection of the SBS4 mutational signature, which may lead to a better understanding of the long-term effects of smoking on lung cancer etiology and inform prevention strategies.